-Search query
-Search result
Showing all 28 items for (author: lico & c)

EMDB-34073: 
Cryo-EM structure of the Serotonin 6 (5-HT6) receptor-DNGs-scFv16 complex
Method: single particle / : Zhao QY, Wang YF, He L, Wang S, Cong Y

PDB-7ys6: 
Cryo-EM structure of the Serotonin 6 (5-HT6) receptor-DNGs-scFv16 complex
Method: single particle / : Zhao QY, Wang YF, He L, Wang S, Cong Y

EMDB-29044: 
Structure of Zanidatamab bound to HER2
Method: single particle / : Worrall LJ, Atkinson CE, Sanches M, Dixit S, Strynadka NCJ
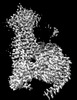
EMDB-32006: 
Cryo-EM structure of Gi coupled Sphingosine 1-phosphate receptor bound with S1P
Method: single particle / : Yu LY, Gan B, Xiao QJ, Ren RB
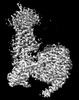
EMDB-32007: 
Cryo-EM structure of Gi coupled Sphingosine 1-phosphate receptor bound with (S)-FTY720-P
Method: single particle / : Yu LY, Gan B, Xiao QJ, Ren RB
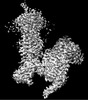
EMDB-32008: 
Cryo-EM structure of Gi coupled Sphingosine 1-phosphate receptor bound with CBP-307
Method: single particle / : Yu LY, Gan B, Xiao QJ, Ren RB

EMDB-32009: 
Cryo-EM structure of Gi coupled Sphingosine 1-phosphate receptor bound with CBP-307
Method: single particle / : Yu LY, Gan B, Xiao QJ, Ren RB

PDB-7vie: 
Cryo-EM structure of Gi coupled Sphingosine 1-phosphate receptor bound with S1P
Method: single particle / : Yu LY, Gan B, Xiao QJ, Ren RB

PDB-7vif: 
Cryo-EM structure of Gi coupled Sphingosine 1-phosphate receptor bound with (S)-FTY720-P
Method: single particle / : Yu LY, Gan B, Xiao QJ, Ren RB

PDB-7vig: 
Cryo-EM structure of Gi coupled Sphingosine 1-phosphate receptor bound with CBP-307
Method: single particle / : Yu LY, Gan B, Xiao QJ, Ren RB

PDB-7vih: 
Cryo-EM structure of Gi coupled Sphingosine 1-phosphate receptor bound with CBP-307
Method: single particle / : Yu LY, Gan B, Xiao QJ, Ren RB

EMDB-10996: 
Structure of Mycobacterium smegmatis HelD protein in complex with RNA polymerase core - State I, primary channel engaged
Method: single particle / : Kouba T, Koval T

EMDB-11004: 
Structure of Mycobacterium smegmatis HelD protein in complex with RNA polymerase core - State II, primary channel engaged and active site interfering
Method: single particle / : Kouba T, Koval T, Krasny L, Dohnalek J

EMDB-11026: 
Structure of Mycobacterium smegmatis HelD protein in complex with RNA polymerase core - State III, primary channel dis-engaged and active site interfering
Method: single particle / : Kouba T, Koval T

PDB-6yxu: 
Structure of Mycobacterium smegmatis HelD protein in complex with RNA polymerase core - State I, primary channel engaged
Method: single particle / : Kouba T, Koval T, Krasny L, Dohnalek J

PDB-6yys: 
Structure of Mycobacterium smegmatis HelD protein in complex with RNA polymerase core - State II, primary channel engaged and active site interfering
Method: single particle / : Kouba T, Koval T, Krasny L, Dohnalek J

PDB-6z11: 
Structure of Mycobacterium smegmatis HelD protein in complex with RNA polymerase core - State III, primary channel dis-engaged and active site interfering
Method: single particle / : Kouba T, Koval T, Krasny L, Dohnalek J

EMDB-4740: 
Atomic structure of potato virus X, the prototype of the Alphaflexiviridae family
Method: helical / : Grinzato A, Kandiah E

PDB-6r7g: 
Atomic structure of potato virus X, the prototype of the Alphaflexiviridae family
Method: helical / : Grinzato A, Kandiah E, Lico C, Betti C, Baschieri S, Zanotti G

PDB-6rvv: 
Structure of left-handed protein cage consisting of 24 eleven-membered ring proteins held together by gold (I) bridges.
Method: single particle / : Malay AD, Miyazaki N, Biela AP, Iwasaki K, Heddle JG

PDB-6rvw: 
Structure of right-handed protein cage consisting of 24 eleven-membered ring proteins held together by gold (I) bridges.
Method: single particle / : Malay AD, Miyazaki N, Biela AP, Iwasaki K, Heddle JG

EMDB-4443: 
Structure of left-handed protein cage consisting of 24 eleven-membered ring proteins held together by gold (I) bridges.
Method: single particle / : Malay AD, Miyazaki N, Biela AP, Iwasaki K, Heddle JG

EMDB-4444: 
Structure of right-handed protein cage consisting of 24 eleven-membered ring proteins held together by gold (I) bridges.
Method: single particle / : Malay AD, Miyazaki N, Biela AP, Iwasaki K, Heddle JG

EMDB-6966: 
Structure of protein cage consisting of 24 eleven-membered ring proteins induced by addition of gold nanoparticle (GNP)
Method: single particle / : Malay AD, Miyazaki N, Biela A, Chakraborti S, Majsterkiewicz K, Kaplan CS, Hochberg GKA, Wu D, Wrobel TP, Benesch JLP, Iwasaki K, Heddle JG, Kelemen P, Vavpetic P, Pelicon P

EMDB-4192: 
Structure of Mycobacterium smegmatis RNA polymerase core
Method: single particle / : Kouba T, Barvik I

PDB-6f6w: 
Structure of Mycobacterium smegmatis RNA polymerase core
Method: single particle / : Kouba T, Barvik I, Krasny L
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search





 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
